The role of mucosal T cell responses in aging, vaccination, and infection
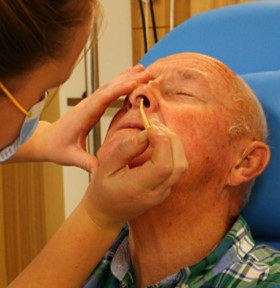 We investigate T cells in the context of aging and respiratory mucosal immunity, focusing on the tissue-resident memory T cells in the nose that protect against airway pathogens1. Through global collaborations and controlled human infection studies (https://www.musicc.org/ and https://www.fluniversal.eu/), we explore how vaccination and infection shape antigen-specific T cell responses in the upper respiratory tract. We have developed and apply minimally invasive sampling methods2,3 and advanced techniques like single-cell RNA sequencing, high-dimensional spectral flow cytometry and machine learning to study these processes in detail4.
We investigate T cells in the context of aging and respiratory mucosal immunity, focusing on the tissue-resident memory T cells in the nose that protect against airway pathogens1. Through global collaborations and controlled human infection studies (https://www.musicc.org/ and https://www.fluniversal.eu/), we explore how vaccination and infection shape antigen-specific T cell responses in the upper respiratory tract. We have developed and apply minimally invasive sampling methods2,3 and advanced techniques like single-cell RNA sequencing, high-dimensional spectral flow cytometry and machine learning to study these processes in detail4.
Organoid-based models to study airway immunity
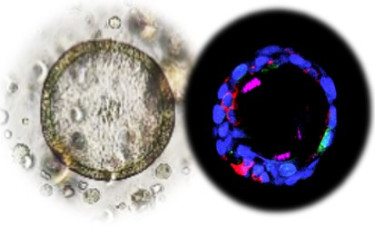 We have developed organoid-based in vitro models derived from primary nasal epithelial cells, collected using minimally-invasive methods, that incorporate autologous immune cells5. These in vitro patient analogues allow mechanistic studies of nasal mucosal immunity, infection responses and epithelial-immune interactions, and provide understanding of how these are influenced by aging, vaccination, and respiratory viral infections. We collaborate within the INNO4VAC consortium (https://www.inno4vac.eu/) to translate in vitro findings to in vivo settings to improve the biological relevance and predictive power of our organoid-based models.
We have developed organoid-based in vitro models derived from primary nasal epithelial cells, collected using minimally-invasive methods, that incorporate autologous immune cells5. These in vitro patient analogues allow mechanistic studies of nasal mucosal immunity, infection responses and epithelial-immune interactions, and provide understanding of how these are influenced by aging, vaccination, and respiratory viral infections. We collaborate within the INNO4VAC consortium (https://www.inno4vac.eu/) to translate in vitro findings to in vivo settings to improve the biological relevance and predictive power of our organoid-based models.
Pediatric respiratory tract infection and host response kinetics
 Our research examines transmission dynamics and infection kinetics in children, focusing on bacterial and viral pathogens. Using high-frequency, child-friendly nasal sampling, we track infection dynamics over time and link these patterns to local immune responses and symptoms6. We also investigate transmission pathways and evaluate immune responses to nasal vaccination, providing insights into pediatric respiratory health and prevention strategies.
Our research examines transmission dynamics and infection kinetics in children, focusing on bacterial and viral pathogens. Using high-frequency, child-friendly nasal sampling, we track infection dynamics over time and link these patterns to local immune responses and symptoms6. We also investigate transmission pathways and evaluate immune responses to nasal vaccination, providing insights into pediatric respiratory health and prevention strategies.
Tracking vaccine responses in lymphoid organs
![]() We use ultrasound-guided lymph node fine needle aspirations to monitor pneumococcal vaccine responses within draining lymph nodes. Our studies assess how these responses that generate immune memory vary by vaccine platform, aging, and HIV infection. Immune profiling includes antigen-specific B and T cells using spectral flow cytometry, enabling comparisons between lymph nodes and peripheral blood to better understand localized and systemic vaccine-induced immunity.
We use ultrasound-guided lymph node fine needle aspirations to monitor pneumococcal vaccine responses within draining lymph nodes. Our studies assess how these responses that generate immune memory vary by vaccine platform, aging, and HIV infection. Immune profiling includes antigen-specific B and T cells using spectral flow cytometry, enabling comparisons between lymph nodes and peripheral blood to better understand localized and systemic vaccine-induced immunity.
High-dimensional profiling of antigen-specific B cells
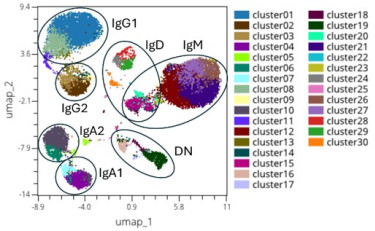 Pneumococcal vaccines show variable effectiveness, underscoring the need to understand cellular immune responses induced by these vaccines. We have developed methods to quantify and characterize B cells, detecting over 25 antigen-specificifities simultaneously, including polysaccharide and other antigens7. By extending beyond antibody responses to B cell immunity, we aim to uncover mechanisms underlying durable protection.
Pneumococcal vaccines show variable effectiveness, underscoring the need to understand cellular immune responses induced by these vaccines. We have developed methods to quantify and characterize B cells, detecting over 25 antigen-specificifities simultaneously, including polysaccharide and other antigens7. By extending beyond antibody responses to B cell immunity, we aim to uncover mechanisms underlying durable protection.
Unveiling SES-associated immune signatures and population diversity
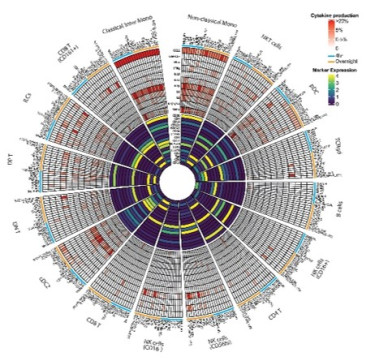 To study socioeconomic status (SES) effects on immunity, we have established a spectral flow cytometry platform profiling over 300 cell surface proteins across immune subsets, but also methods to comprehensively characterize the functional capacity of the innate immune system8. These tools allow us to characterize immune variation in SES-stratified populations but also map how aging changes across population. We also investigate how SES influences differential responses to infection at the airway mucosa9, providing insights into population diversity and health disparities.
To study socioeconomic status (SES) effects on immunity, we have established a spectral flow cytometry platform profiling over 300 cell surface proteins across immune subsets, but also methods to comprehensively characterize the functional capacity of the innate immune system8. These tools allow us to characterize immune variation in SES-stratified populations but also map how aging changes across population. We also investigate how SES influences differential responses to infection at the airway mucosa9, providing insights into population diversity and health disparities.
1 Roukens, A. H. E. et al. Prolonged activation of nasal immune cell populations and development of tissue-resident SARS-CoV-2-specific CD8(+) T cell responses following COVID-19. Nat Immunol 23, 23-32, (2022).
2 Jochems, S. P. et al. Inflammation induced by influenza virus impairs human innate immune control of pneumococcus. Nat Immunol 19, 1299-1308, (2018).
3 Jochems, S. P. et al. Novel Analysis of Immune Cells from Nasal Microbiopsy Demonstrates Reliable, Reproducible Data for Immune Populations, and Superior Cytokine Detection Compared to Nasal Wash. PLoS One 12, e0169805, (2017).
4 Mbow, M. et al. Immune responses to SARS-CoV-2 in sub-Saharan Africa and western Europe: a retrospective, population-based, cross-sectional study. Lancet Microbe 6, (2025).
5 King, L.A. et al. Protocol for the generation of a human-derived 3D nasal epithelial model and the induction of tissue-resident memory-like T cells in an in vitro co-culture system. Submitted.
6 Hoving, D. et al. High-resolution characterization of nasal microbial dynamics in young children. medRxiv, 2025.2008.2022.25334231, (2025).
7 Hoving, D. et al. Combinatorial multimer staining and spectral flow cytometry facilitate quantification and characterization of polysaccharide-specific B cell immunity. Commun Biol 6, 1095, (2023).
8 Huisman, W. et al. Immuno-functionomics reveals geographical variation and a role for TLR8 in mRNA vaccine responses. iScience 28, 113839, (2025).
9 van Dorst, M. et al. Differences in Bacterial Colonization and Mucosal Responses Between High and Low SES Children in Indonesia. Pediatr Infect Dis J 41, 496-506, (2022).

